Dr. Marc Fouet, Postdoctoral Researcher
University of California, Berkeley
Background
The Rine lab at the University of California, Berkeley is working towards understanding mechanisms underlying establishment, maintenance, and epigenetic inheritance of gene silencing in yeast. The lab has developed a genetic strategy to capture transient losses of gene silencing of heterochromatin in S. cerevisiae, and translating these dynamic processes as a permanent modification of fluorescence expression. By using the CRASH (cre-reported altered states of heterochromatin) reporter, red fluorescent yeast cells become green once gene silencing by Sir proteins has been lost.1 Monitoring fluorescence of yeast colonies can be used to describe dynamic epigenetic phenomena quantitatively. Recently, the Rine lab began using high-throughput yeast aging analysis (HYAA) chips2 with high-resolution time-lapse microscopy to monitor the relative life span of yeast and understand better the relationship between gene silencing and aging.3 Dr. Marc Fouet, a postdoctoral researcher in the Rine lab, is working on solutions to detect events leading to a lower signal of fluorescence and on the microfluidic process intensification to image an increasing number of single cells.

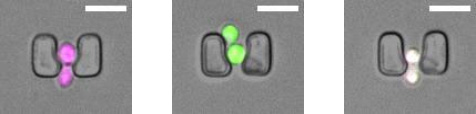
Figure 1 is an image from a chamber with cells carrying the CRASH (cre-reported altered states of heterochromatin) reporter that induces a permananet and hertibale switch from expressing red fluorescent protein (RFP) to expressing green fluorescent protein (GFP) when loss of silencing of the auxiliary mating-type locus HMLα2 occurs.1
Challenge
Rare transcription events within heterochromatin occur in approximately 1/1000 cell divisions and cells divide every 90 minutes. In order to capture these rare events, 40 images are taken of 5,000 yeast cell traps for each sample. Images are captured every 5 to 10 minutes to measure cell division and fluorescent protein levels. Phototoxicity and photobleaching should be minimized, and resolution is increasingly important when machine learning algorithms are used to segment fluorescence data.
The bigger our field of view and the lower the exposure times are, the more cells we can image overall as less images are needed and the acquisition is faster [using the Prime 95B Scientific CMOS camera].
Solution
The Prime 95B Scientific CMOS (sCMOS) camera has a 95% quantum efficicency (QE) and can detect low levels of fluorescent proteins in yeast. The high sensitivity of the Prime 95B camera allowed Marc to lower the exposure time to reduce phototoxicity and photobleaching.
The sensitivity and large field of view reduces acquisition time of the microfluidic chip. In addition to faster exposure times, the large field of view (18 mm diagonal) reduces the amount of images needed per chip and decreases acquisition times.
The Prime 95B camera is a very versatile camera and is useful in multiple applications in the Rine group. In addition to time lapse imaging, the Prime 95B camera can acquire full frame images at 82 fps in the 12-bit mode. This is fast enough to capture rapid events that occur frequently in microfluidic chips, for example, loading of a cell trap.
References
- Dodson AE, Rine J. Heritable capture of heterochromatin dynamics in Saccharomyces cerevisiae. Elife (2015), doi: 10.7554/eLife.05007.
- Jo MC, Liu W, Gu L, Dang W, Qin L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc Natl Acad Sci U S A (2015), doi: 10.1073/pnas.1510328112.
- Schlissel G, Krzyzanowski MK, Caudron F, Barral Y, Rine J. Aggregation of the Whi3 protein, not loss of heterochromatin, causes sterility in old yeast cells. Science (2017), doi: 10.1126/science.aaj2103.

