Introduction
Neurons in the brain communicate with chemical and electricity signals. Neuroscience has developed numerous probes and electrodes in order to detect this activity, and try to figure out what the neurons in each brain area do. This can be expensive and time-consuming. Researchers have had the ability to suppress or enhance the expression of genes for many years. However, the process of initiating this change takes hours, days or even months with traditional genetic techniques, rendering it difficult to measure or even observe the resulting neurological change. Alternatively, electrical stimulation offers some degree of stimulation or suppression of neurons on much faster timescales and can be targeted to a given region. However, electrical stimulation is unable to specifically target individual types of cells.
What if brain activity could be controlled with light? The specialized photoreceptor cells in the eyes are a type of neuron that responds to light and converts the light into a signal, which is what gives us sight. If we could give this ability to react to light to other neurons, it would represent a huge jump in neuroscience, as experiments would take milliseconds and could be targeted to any neuron in the brain. This is the principle of optogenetics, a combination of optical and genetic techniques allowing the firing or suppression of neurons with direct light stimulation.
Theory Of Optogenetics
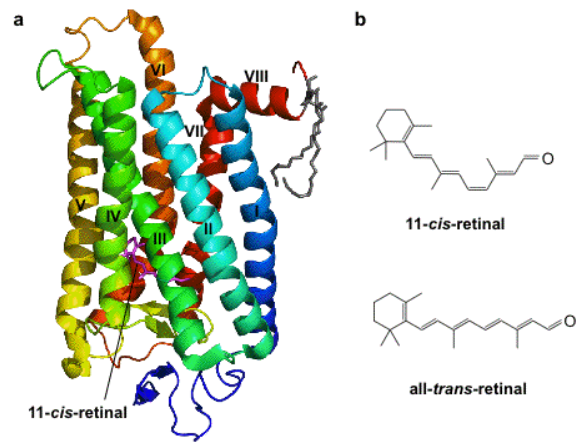
In the eyes of mammals, there are rod and cone cells, which detect light. In our rod cells is a special protein called rhodopsin, which is essential for our ability to see. Rhodopsin is made up of two components, a membrane protein (part of a protein family called opsins) and a light-sensitive chemical (called retinal, a form of vitamin A), as seen in Fig.1. In the rod cells of the eye, when rhodopsin is exposed to light it causes retinal to change shape (depending on the color/wavelength of the light), which pushes the opsin protein and starts a signal to the brain which results in images formed and the perception of light. If neurons in the brain could be encouraged to express these opsins, they would be sensitive to light.
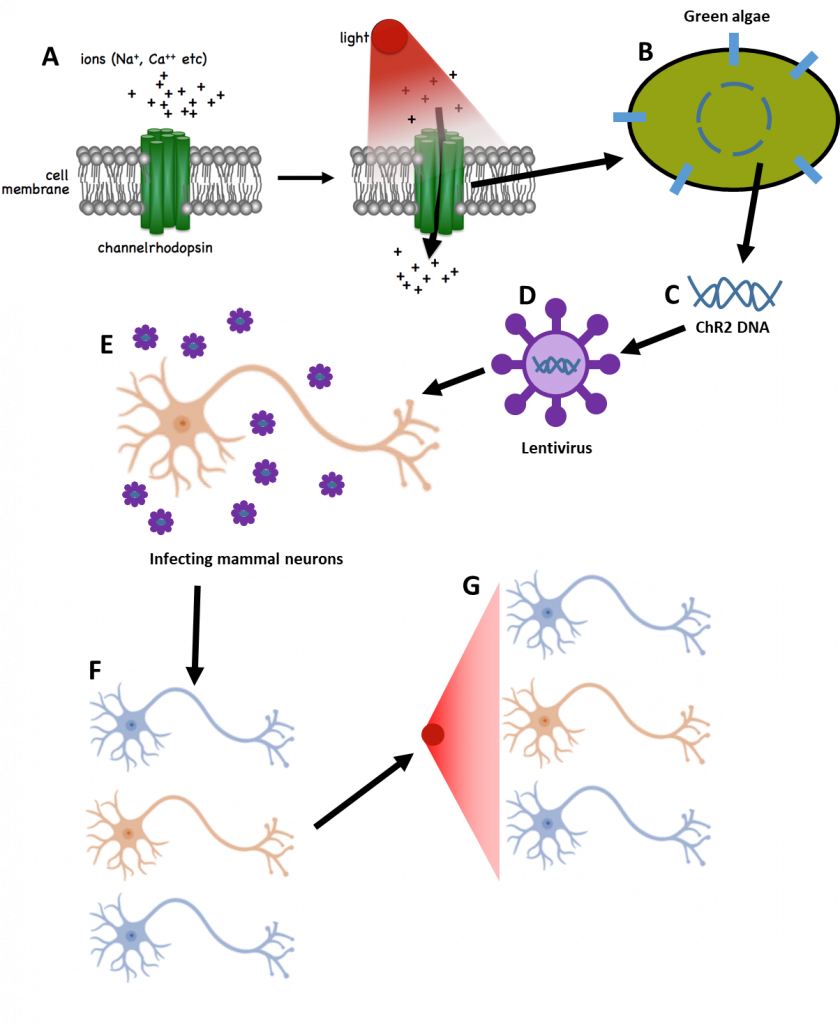
Rhodopsins have also been discovered in bacteria and other microbial organisms, with researchers identifying bacteriorhodopsin, halorhodopsin, and channelrhodopsin. In 2005, Boyden and colleagues took a special interest in a channelrhodopsin-2 (ChR2) from green algae (Fig.2A-B), which reacted to short (50 µs) blue flashes of light by opening an ion channel in the cell membrane (Boyden et al. 2005). By extracting the gene for ChR2 from the algae (Fig.2B-C) and adding it into the genetic material of mammal neurons, neurons could be produced that had their own ChR2 and would react to blue light by opening ion channels, activating the cell. This was done by taking the ChR2 gene from algae and inserting it into a special type of virus called a lentivirus (Fig.2D). By infecting neurons grown in the lab with this lentivirus (Fig.2E), the ChR2 gene was introduced to the neurons by the virus and the neurons reacted to blue light with activity (Fig.2F-G). This marks one of the first times that neurons could be turned on with just light, namely photo‑switchable neurons. This is the core concept of optogenetics, where genetic techniques are used to create light-sensitive cells for study. This process is outlined in Fig.2.
Viruses are typically used in optogenetics to introduce genetic material to cells. Viruses provide both fast and versatile insertion, with a high proportion of target cells being infected, providing robust expression. There are a number of ways of ensuring that the viruses infect the specific cell type that needs to be activated via light, including using specific promoters, careful targeting of the injection of the virus, and precisely targeting the light delivery. However, most viral vectors are restricted in the amount of genetic information they can carry. An alternative way to introduce the opsins to mammalian cells is to use genetic ‘knock-in’ techniques to raise animals that express the proteins as part of their own genetic code, which avoids the limited carrying ability of viruses. However, introducing opsins in this way requires more time, effort and cost (Fenno, Yizhar and Deisseroth, 2011).
Once in place, rhodopsins allow manipulation of neurons on timescales of milliseconds rather than months. The result is a technique that can be used to both study the role of neurons and control the behavior of an organism, either in vivo or in vitro. In vivo, equipment to control neuron behavior and monitor neuron activity can be implanted into the brain of any freely-moving animal from a fruit fly to a primate. In vitro, optical stimulation can be combined with optical and electrophysiological techniques to observe the results on the cellular level.
Optogenetics not only provides valuable insight into the normal functioning of brain tissues but also into neurological conditions such as Parkinson’s disease, Alzheimer’s disease and epilepsy. And in the future, optically controllable cells may themselves be used to treat disorders.
Optogenetic Control
There are three goals for optogenetic studies, each of which requires a different class of opsin.
- Stimulating the firing of a neuron
- Suppressing the firing of a neuron
- Biasing the response of a neuron to make spontaneous firing more likely
Opsins that cause the neuron to fire are the most common, and there are dozens of choices that can be activated with a broad range of wavelengths of light, also with a broad range of response times from milliseconds to tens of minutes. A summary of commonly used opsins can be found in Table 1.
Table 1: Common opsins for neural activity modulation, adapted from Mahmoudi, Veladi, and Pakdel (2017).
| Opsin name | Action | Opsin type | Wavelength (nm) |
| ChR2 | Activator | Na+/K+ channel | 400-500 |
| NpHR | Inhibitor | Cl– pump | 550-620 |
| ReaChR | Activator | Na+/K+ channel | 590-630 |
| Chrimson | Activator | Na+/K+ channel | 600 |
| SFO | Activator | Na+/K+ channel | 450-590 |
| VChR1 | Activator | Na+/K+ channel | 500-550 |
| iC1C2 | Inhibitor | Cl– channel | 450-500 |
Opsins that inhibit neuronal activity with light are rarer than those that excite neurons, but several inhibitory opsins have been identified or artificially engineered (Wietek et al. 2014; Berndt et al. 2014).
A more subtle approach is to avoid directly causing the neuron to fire with a given trigger, and instead to put the neuron into an ‘excited state’ where firing becomes more likely. This can be achieved using a ‘step-function opsin’. These further have the advantage that the neuron remains in the excited state long after the light is deactivated, meaning the optical fiber to supply the pulse can then be disconnected, allowing the animal free movement. Developments in step-function opsins have led to neurons that can exhibit up to 30 minutes of biased activity after a single 10ms blue light pulse, which can also be deactivated with a pulse of yellow light (Yizhar et al. 2011).
Optogenetics Observations
While in vivo measurements can study the macroscopic behavior of an organism under the optogenetic influence, in vitro experiments can provide control and observation with single-cell resolution. For in vitro experiments, neuronal cells are typically prepared in culture. Cells are simply illuminated with either filtered light from mercury arc lamps or LEDs within a microscope setup, though more advanced lighting setups to very specifically target subregions of the sample are available.
Illumination in vivo is a little more complicated as typically the animal still needs to be able to move and behave as normal, but typically involves the implantation of a flexible optical fiber into the brain, with laser or high-power LED light used for illumination. However, this approach is fairly invasive and still restricts movement. In 2013, Bruchas and Rogers developed a wireless, ultrathin needle-based system for control and observation that can be injected deep into soft tissues allowing a greater degree of freedom for the animal (T. -i. Kim et al. 2013). This device uses 8.5 μm thick micro-LEDs, a microelectrode, an integrated photodetector and a temperature sensor to both optically stimulate and monitor the brain of a moving animal.
Both in vivo and in vitro studies must occasionally image deep into thick tissues. One method to achieve this is two-photon microscopy with a pulsed laser. This technique allows simultaneous control and observation, with very good spatial and temporal resolution (Andrasfalvy et al. 2010). In vivo, 2-photon calcium imaging can allow deeper penetration into living tissues to observe the influence of optogenetic stimulation in real-time. However, these techniques require the animal to be stationary, such as on a fixed treadmill (C. K. Kim et al. 2017).
Optogenetics Discoveries
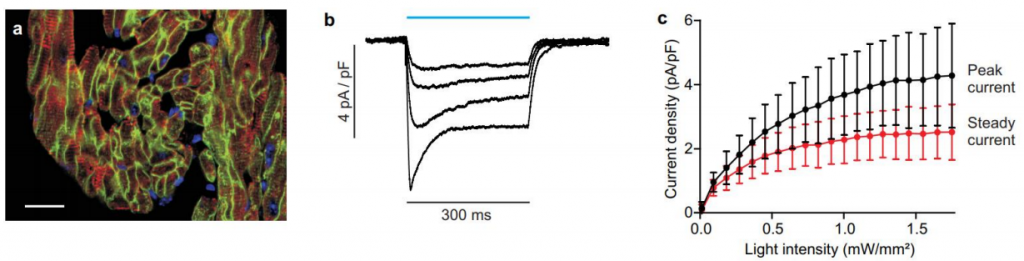
To observe the results of optogenetic control on a microscopic level as well as the macroscopic ‘behavioral’ level, electrophysiological techniques are frequently used in parallel, often with control and observation hardware built into a single unit. For in vitro studies, microscopy techniques are also possible, such as studying the beating of heart muscle cells that are under optogenetic control (Bruegmann et al. 2010). This allowed the researchers to stimulate, study and control heart muscles without the toxic effects of electrical stimulation, providing a mechanism for investigating pacemaking. Fluorescence imaging shown in Fig.3A confirms the cell specificity of the light-sensitive proteins, and Fig.3B-C confirm the electrical response of the heart tissues to light.
A recent in vivo study in mice used optogenetics to discover pathways of the brain responsible for ‘reward-seeking’ behavior (C. K. Kim et al. 2017). Through the optogenetic enhancement of these pathways, the researchers caused a mouse to be less likely to press a lever that had a likelihood of delivering either a morsel of food or a mild shock. This may pave the way for treatments of psychiatric illnesses that heighten reward-seeking such as substance abuse, or suppress it, such as depression. By confining the mouse to a treadmill, researchers were able to mount apparatus to its cranium in order to perform two-photon calcium imaging during behavior, confirming the microscopic response.
One major consideration for any behavior study is to ensure that the experimental apparatus, along with any genetic changes to the organism, don’t influence behavior. For optogenetics, this often requires running parallel experiments with and without cranially-implanted apparatuses, and with and without genetic modification. This is on top of any control groups necessary to isolate the effect of the optogenetic stimulation. Additionally, to prepare a sample for optogenetic control or observation with cell-type specificity often requires difficult and advanced techniques, and requires careful validation, especially check avoidance non-targeted populations.
Cameras For Optogenetics
The demands for camera imaging in optogenetics are very broad, from the macroscopic observation of organisms as large as primates to the high-speed, high-magnification microscopic observation of individual cells. In vivo studies typically observe animal behavior as a key experimental outcome. Video or time-lapse imaging of the animal freely moving within its environment is often used.
On the microscopic scale, fluorescence imaging of the targeted cells can be a vital tool in verifying the accuracy of cell-type targeting methods. For in vitro experiments, the tools of optogenetics often go hand-in-hand with those of electrophysiology in general such as patch-clamping and calcium imaging, which is covered in greater detail in the application notes on electrophysiology and calcium imaging, also available in this section of our website.
Summary
For microscopists with powerful LED and laser illumination on hand and the ability to use electrophysiological techniques in parallel, the ability to control neuronal cells with millisecond time resolution and cell-type specificity allow an unprecedented degree of insight into the cellular-level behavior of the brain.
Optogenetics is an excellent tool in the arsenal of neuroscientists, and the wealth of research using these techniques in everything from small-scale in vitro experiments to human clinical trials of medical procedures is a testament to its potential.
Download As PDF
References
Andrasfalvy, Bertalan K, Boris V Zemelman, Jianyong Tang, and Alipasha Vaziri. 2010. “Two-Photon Single-Cell Optogenetic Control of Neuronal Activity by Sculpted Light.” Proceedings of the National Academy of Sciences 107 (26). National Acad Sciences:11981–86.
Berndt, Andre, Soo Yeun Lee, Charu Ramakrishnan, and Karl Deisseroth. 2014. “Structure-Guided Transformation of Channelrhodopsin into a Light-Activated Chloride Channel.” Science 344 (6182). American Association for the Advancement of Science:420–24.
Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci, 8(9), 1263-8.
Bruegmann, Tobias, Daniela Malan, Michael Hesse, Thomas Beiert, Christopher J Fuegemann, Bernd K Fleischmann, and Philipp Sasse. 2010. “Optogenetic Control of Heart Muscle in Vitro and in Vivo.” Nature Methods 7 (11):897– 900. https://doi.org/10.1038/nmeth.1512.
Deisseroth, Karl. 2015. “Optogenetics: 10 Years of Microbial Opsins in Neuroscience.” Nature Neuroscience 18 (9):1213–25. https://doi.org/10.1038/nn.4091.
Fenno, Lief, Ofer Yizhar, and Karl Deisseroth. 2011. “The Development and Application of Optogenetics.” Annual Review of Neuroscience 34 (1):389–412. https://doi.org/10.1146/annurev-neuro-061010-113817.
Kim, Christina K., Li Ye, Joshua H. Jennings, Nandini Pichamoorthy, Daniel D. Tang, Ai Chi W. Yoo, Charu Ramakrishnan, and Karl Deisseroth. 2017. “Molecular and Circuit-Dynamical Identification of Top-Down Neural Mechanisms for Restraint of Reward Seeking.” Cell 170 (5):1013–1027.e14. https://doi.org/10.1016/j.cell.2017.07.020.
Kim, T.-i., J. G. McCall, Y. H. Jung, X. Huang, E. R. Siuda, Y. Li, J. Song, et al. 2013. “Injectable, Cellular-Scale Optoelectronics with Applications for Wireless Optogenetics.” Science 340 (6129):211–16. https://doi.org/10.1126/science.1232437.
Mahmoudi, Parisa, Hadi Veladi, and Firooz G Pakdel. 2017. “Optogenetics, Tools and Applications in Neurobiology.” Journal of Medical Signals and Sensors 7 (2). Medknow Publications:71.
Nagata T., Koyanagi M and Terakita A. (2010) Molecular Evolution and Functional Diversity of Opsin-Based Photopigments, Osaka City University, http://photobiology.info/Terakita.html
Wietek, Jonas, J Simon Wiegert, Nona Adeishvili, Franziska Schneider, Hiroshi Watanabe, Satoshi P Tsunoda, Arend Vogt, Marcus Elstner, Thomas G Oertner, and Peter Hegemann. 2014. “Conversion of Channelrhodopsin into a Light-Gated Chloride Channel.” Science 344 (6182). American Association for the Advancement of Science:409– 12.
Yizhar, Ofer, Lief E Fenno, Matthias Prigge, Franziska Schneider, Thomas J Davidson, Daniel J O’Shea, Vikaas S Sohal, et al. 2011. “Neocortical Excitation/inhibition Balance in Information Processing and Social Dysfunction.” Nature 477 (7363). Nature Research:171–78.
