Introduction
Current methods used to construct 3D optical images are limited in the maximum thickness of certain samples. Confocal microscopy (in combination with two-photon approaches) is limited to ~1 mm depth, meaning that larger samples would have to be further sectioned and would not remain intact. Optical coherence tomography (OCT) can image to greater depths of ~2-3 mm, but this is still too little to image large biological samples such as whole embryos.
Tomography is the term for imaging by sections, and is used in all walks of science to build up 3D volumes out of 2D slice images. The principle of tomography can be seen in Fig.1.
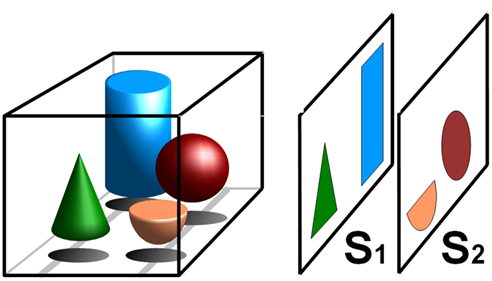
Current forms of 3D optical microscopy use section tomography, where optical sections are generated through a sample by focusing on various 2D planes and minimizing noise or out-of-focus signal from the surrounding planes. This raw data is then mapped into a 3D object, as each discrete image relates to a section of that volume, known as a voxel (a 3D pixel/volumetric pixel). This approach is optimized for taking very sharp, clean and submicron resolution images of small specimens, such as cells and tissues, but is unsuitable for large volumetric 3D samples such as whole embryos/organisms. Light sheet techniques such as single-plane illumination microscopy (SPIM) can image large specimens, but is limited by the need for localized fluorescent excitation (Sharpe et al. 2009).
The alternative is projection tomography. In this technique, the raw data is taken from many angles around the sample and mathematically transformed to recreate the original 3D object. A good example of section tomography is the medical computerized tomography or CT scanner, which uses x-rays that can easily pass through biological tissue. This kind of projection tomography can also be performed with optical wavelengths (ultraviolet, visible light and infrared), in order to image as deep as 10 cm into live biological samples (increased depth resulting in decreased resolution). This is known as optical projection tomography (OPT).
Optical Projection Tomography
OPT is a relatively recent technique, developed in 2002 by Sharpe and colleagues due to current 3D optical methods being limited on the maximum thickness of 3D samples that could be imaged. OPT allows for high-resolution 3D images of fluorescent and non-fluorescent biological samples with a thickness of up to 15 mm.
OPT can be used in two main modes, transmission or emission. Transmission modes require light to pass through a sample to obtain structural data about the sample, but as most biological samples – such as cells, tissues, and whole organisms – are opaque to visible light, these samples often need to be artificially/chemically cleared in order for the light to pass through.
Emission modes rely on sample fluorescence, samples are exposed to excitation light and emitted fluorescence can be used to obtain functional information about the sample. This emission OPT is also known as fluorescence optical projection tomography or F-OPT.
OPT Microscopy
Sharpe and colleagues paired OPT to a microscope, allowing both standard fluorescent and brightfield imaging. The light path found in OPT microscopy can be seen in Fig.2.
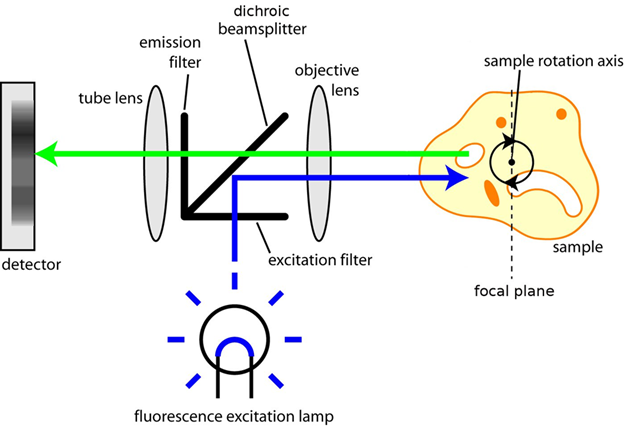
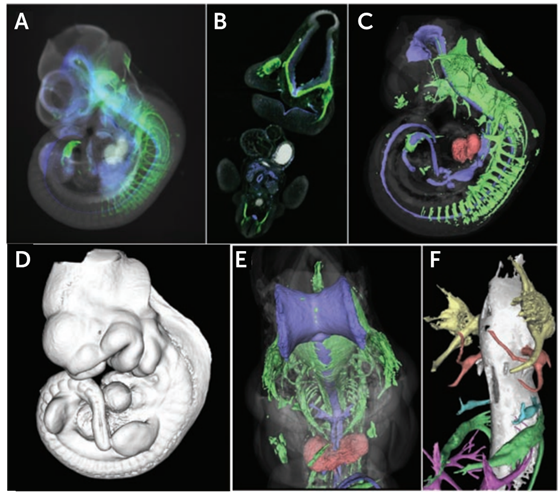
By clearing a mouse embryo and imaging with transmission OPT as the sample rotated, the virtual sections of an embryo were independently reconstructed using a back-projection algorithm (details on filtered back-projection can be found in McGinty et al. (2008). This algorithm spreads image information across a blank image from the projection rows of obtained voxels, resulting in a high-resolution representation of the sections through the specimen, and a complete 3D image of a whole mouse embryo (seen in Fig.3).
F-OPT allows for virtual sectioning of a large sample and reconstruction of a high-resolution 3D data set in any orientation. The ability to recreate very fine structures such as developing nerve fibers (see Fig.3F) in 3D demonstrates the power of OPT to align serial sections with extremely high accuracy.
Advantages and Disadvantages Of OPT
The main advantage of OPT is the ability to image fluorescent tags, across different tissues or regions expressing particular proteins, mRNAs or other molecules of interest. Current instrumentation allows for visualization in several UV or visible light channels, allowing simultaneous imaging of several markers as well as generation of 3D morphology in a single scan session. This combination of molecular and morphological imaging makes OPT a very powerful technique for tracking morphological and functional changes in developmental biology, as not many techniques can image an entire embryo.
OPT has also successfully been combined with other advanced fluorescent techniques such as fluorescence lifetime imaging (FLIM), creating FLIM-OPT image systems that can construct fluorescence lifetimes throughout large 3D samples (McGinty et al. 2008).
The main drawback of OPT is sample preparation, transmission OPT requires chemical clearing of samples, which is a time‑consuming process and can create processing artifacts that affect sample morphology. In addition, processing and imaging are time-consuming when compared to other techniques such as μCT, and the reliance of OPT on lenses to focus images limits the depth of focus and the maximum size of tissue that can be imaged (Sharpe et al. 2009).
Cameras for OPT
As OPT is optimized for large samples, a camera with a large field of view (FOV) would allow for easier capture of large samples without the need for many 100-1000s of images. In addition, as samples in OPT are typically rotated in order to gather images from many angles, synchronizing a high-speed camera with a high-speed sample rotation would speed up acquisition. A camera with advanced triggering options would also assist with imaging across multiple wavelengths for different fluorophores, while synchronizing the camera exposure and the light source(s) to the sample rotation, allowing for lower exposures, faster acquisitions, and less photobleaching.
sCMOS cameras feature these high speeds, large FOVs, and detailed triggering options, making them a good fit for OPT microscopy applications.
Summary
OPT is a powerful 3D imaging technology when working with large samples, and has been adopted by the developmental biology community for imaging of whole embryos and developing organs. Despite the relatively recent development of OPT, it has found many uses in studying embryogenesis, disease development and more in whole embryo samples. While other techniques such as confocal are limited in their 3D imaging depth, OPT allows for high-resolution imaging down to more than 15 mm through a sample, making it a versatile imaging platform.
References
McGinty J, Tahir KB, Laine R, Talbot C.B., Dunsby C., Neil M.A.A., Quintana L., Swoger J., Sharpe J. and French P.M.W. Fluorescence lifetime optical projection tomography. J Biophotonics. 2008;1(5):390‐394. doi:10.1002/jbio.200810044
Sharpe J., Ahlgren U., Perry P., Hill B., Ross A, Hecksher J.S, Baldock R., Davidson D. (2002) Optical Projection Tomography as a Tool for 3D Microscopy and Gene Expression Studies, Science: 541-545.
Sharpe J. (2009) Optical Projection Tomography. In: Sensen C.W., Hallgrímsson B. (eds) Advanced Imaging in Biology and Medicine. Springer, Berlin, Heidelberg
