Dr. Yandong Yin, Postdoctoral Fellow
Prof. Eli Rothenberg, PhD, Associate Professor
New York University, School of Medicine
Background
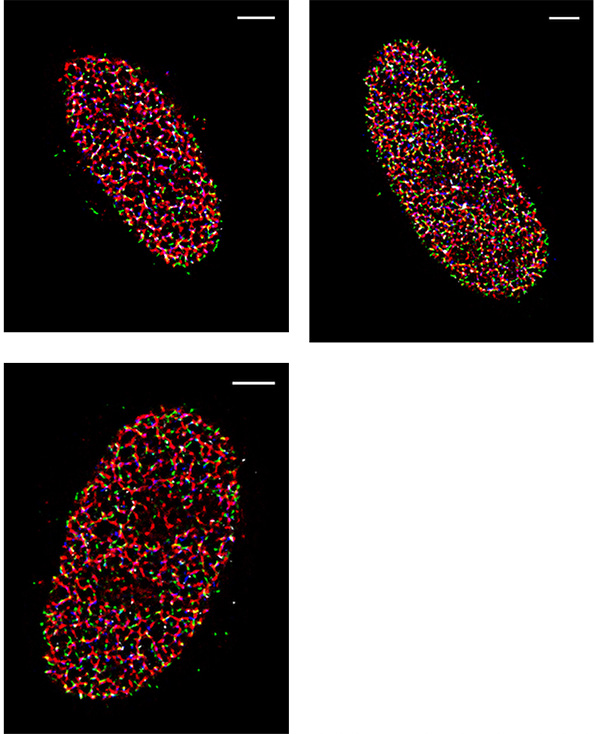
The laboratory of Dr. Rothenberg at the New York University School of Medicine focuses on new optical methods to study biological molecules and processes at real time and nanometer scale. The Rothenberg research team studies the mechanisms of enzymes and proteins that participate in repair of DNA damage leading to cancer, and develops new imaging methods that will enable them to visualize the behavior of individual biological molecules. STORM Microscopy is used to localize and track DNA as it replicates in the cell. “We try to look at the nucleus of cancer cells as they are replicating the DNA. The DNA and proteins involved in DNA replication are labelled so we can understand what is going on when replication happens,” Yandong Yin, PhD. Postdoctoral fellow states.
Exposure time: 30 ms
Magnification: 150 times (the configured pixel size is ~ 73 nm)
Reconstruction Algorithm Used: Maximum Likelihood Estimation (MLE) method for single PSF fitting
Challenge
One of the challenges of imaging replicating DNA is that inside the nucleus of the cell there are many labeled components crowded together, as well as very small components that need to be clearly resolved. To determine how each component is organized spatially, the lab often performs STORM imaging using three or four colors sequentially, which makes resolution, sensitivity, and localization accuracy a great concern. “The DNA replication fork is very small. We can’t image it without super resolution,” says Yin. The laboratory calibrates their STORM post-processing conditions based on the variances of each pixel in the chip of the camera, correcting for any major variations, in order to better fit the point spread function of each fluorophore. Because of this, pixel to pixel variations, like those seen in patterned noise on CMOS cameras becomes a major problem.
If you have a shorter exposure time, you can track faster kinetics. More sensitivity and shorter exposure times with the Prime 95B [Scientific CMOS camera] allow you to image faster and track kinetics better.
Solution
The move away from EMCCD technology to the Prime 95B back-thinned CMOS was made easier because of the improved sensitivity in the detection of low-emission fluorophores, and the reduction in pattern noise when compared to other CMOS cameras. “For single-molecule localization imaging, the most important thing is the reconstruction process in how we fit each single-molecule point spread function into its centroid coordinate,” Yin noted on his use of the Prime 95B. The post-processing of images collected with the Prime 95B are made significantly easier because of the reduction in pixel to pixel pattern noise, which makes the localization and reconstruction of fluorophores of multiple colors easier to do. Yin continued, “The variance for each pixel is much smaller than what is reported on other sCMOS cameras. We have found that more than 90% of the pixels fall within a very tight noise distribution.”
Additionally, the large field of view coupled with the improved sensitivity allows the team to get an image that contains useful information more often. Since they often work with samples that are dark at the start of acquisition, it was an issue that they couldn’t guarantee seeing something with the smaller field of view of an EMCCD. Yin says “Previously we used an EMCCD, but the EMCCD has a smaller chip. With a bigger chip we can see multiple cells simultaneously.”

