Introduction To Super-Resolution
Light microscopy techniques have been vital to our understanding of biological structures in cells and tissues since their invention in the late 16th century. However, the resolution of conventional light microscopy techniques is limited by the diffraction limit of light which prevents structures smaller than approximately ~300 nm from being resolved.
This limit has since been broken by various super-resolution techniques which has led to a greater understanding of biological structures than conventional light microscopy allows. In some cases, structures can now be resolved down to single nanometers.
Existing super-resolution techniques can be broadly classified into three major groups:
- Single molecule localization techniques liked STORM, PALM and DNA-PAINT
- Sculpted light techniques like SIM, STED and RESOLFT
- Computational techniques like SRRF, SOFI and HAWK
More can be read about these techniques in our super-resolution resources.
Expansion microscopy (ExM) represents a new type of super-resolution technique that might be defined instead as a sample preparation technique.
What Is Expansion Microscopy?
Rather than using optics or photophysics to separate biological structures from one another, ExM takes the opposite approach and physically enlarges the sample to pull structures further apart.
This is achieved by embedding the sample in a hydrogel which, when expanded, expands the sample with it. The result is a sample where biological structures are physically further apart than before. These structures can then be resolved even with conventional microscopy techniques.
As an example, assuming a microscope with a diffraction limit of 300 nm and a 4.5x expanded sample, the effective resolution would now be ~300 nm/4.5 which is ~70 nm.
No complex optics or photophysics are necessary to get super-resolved images with ExM as it’s all achieved through the sample. It can be performed on almost any standard microscope system with no modification needed, making it relatively simple for any typical microscopy lab to implement.
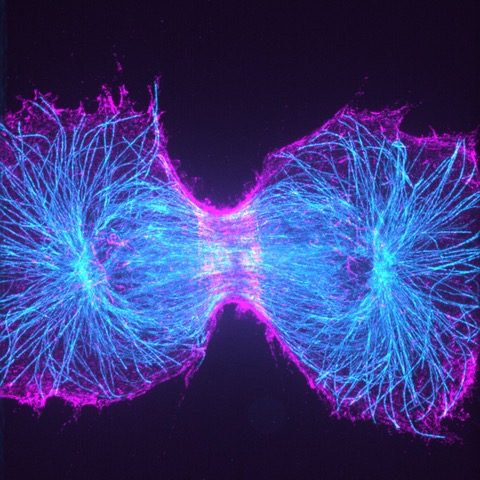
Read more about this work by Prof. Helge Ewers here
How Does Expansion Microscopy Work?
Since the original paper describing the method by Chen et al. (2015), many additional methods and adaptations have been published. The original method for separating proteins will first be summarized and some of the adaptations discussed later.
To give this some context, a popular method of protein staining is antibody staining where a primary antibody binds the protein of interest and then a secondary antibody with a fluorophore binds to the primary antibody, labeling it. The first step in ExM is similar but instead of a fluorophore, the secondary antibody has an oligonucleotide attached that is capable of anchoring to the expansion gel.
Following this sample labeling step, the method proceeds as so:
- The sample is infused with and embedded in a polyelectrolyte hydrogel
- The oligonucleotide bound to the protein of interest anchors itself to the gel, effectively acting as a surrogate for the position of the protein of interest
- Once in the gel, the sample is put into a protease digestion solution. The reason for this is that expansion isn’t possible in native tissue because the tissue holds itself together and prevents expansion. The digestion solution breaks down the proteins and allows expansion
NOTE – It doesn’t matter if the protein of interest gets destroyed in the digestion step, the oligonucleotide has captured its position in the gel - Following digestion, the sample is washed with pure water which pulls out all of the salt. This causes the sample to physically get bigger until it hits a plateau defined by the gel chemistry. In the original protocol this was 4.5x.
- A FISH (fluorescence in situ hybridization) probe or similar fluorophore can then be put into the sample to label the oligonucleotide locations
- Sample imaged on any conventional microscope
ProExM Simplifies The Original Method
Protein-retention expansion microscopy (ProExM) builds on the original method by removing the oligonucleotide synthesis step which added considerable complexity and effort to the initial labeling. It proceeds as so:
- Before embedding in the gel the sample is labeled using typical protein labeling techniques (e.g. antibodies or genetically encoded fluorophores) to stain the proteins of interest
- The sample is infused with a modified gel that allows all proteins to be anchored to it
- Protease digestion is still necessary but applied at a concentration that leaves the protein labels mostly intact.
NOTE – One notable exception are cyanine dyes which get destroyed by the process. - The sample is washed and expanded as in the original method
- Sample imaged on any conventional microscope
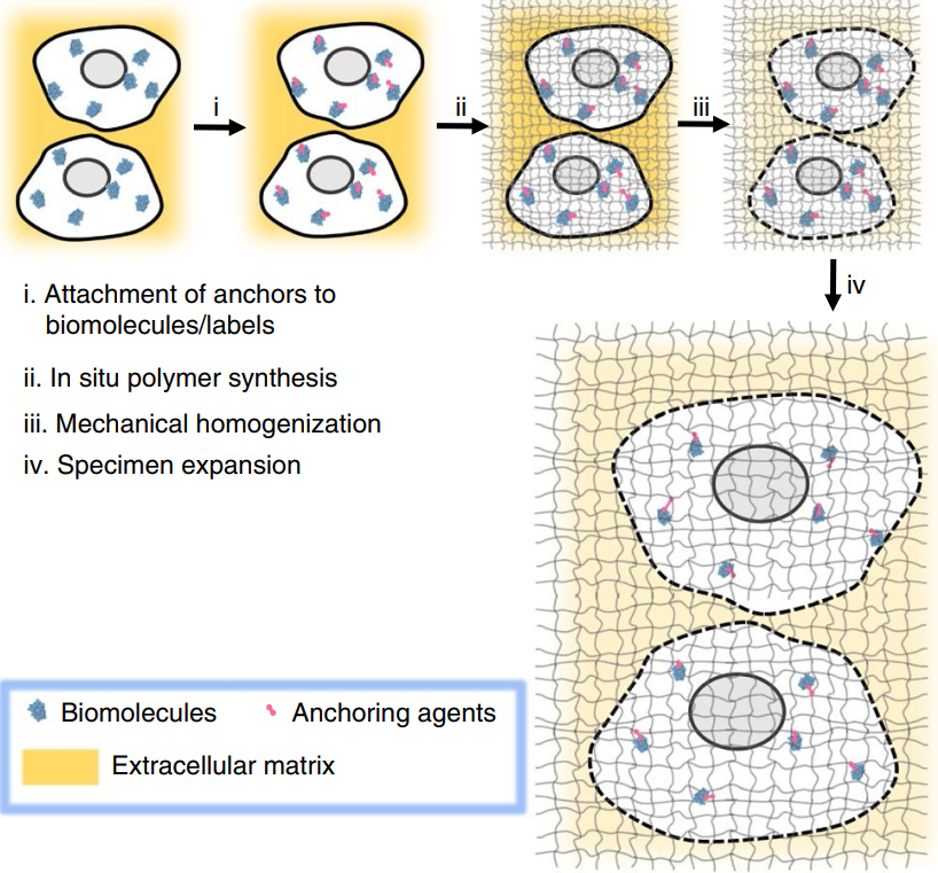
Adapted from Wassie et al. (2018).
Why Use Expansion Microscopy?
ExM has a lot of advantages compared to existing super-resolution techniques, not least providing higher resolution images than many of them. To summarize:
- Expansion is isotropic so axial resolution is improved by the same factor as lateral resolution
- No specific fluorophores are needed, ExM can be performed with most standard fluorophores
- Easy to stain the sample with many different fluorophores for multi-color imaging
- No specific microscope or objective is needed
- Low magnification objectives can be used for large samples and long working distance objectives can be used for deep imaging
- No image reconstruction is needed
- Digestion and expansion clears the sample, making it almost entirely water. This eliminates scattering, improves signal to noise ratio and results in a very clean image
- ExM can be combined with other microscopy techniques like light sheet microscopy or spinning disk confocal to perform three-dimensional imaging and allowing these techniques to become super-resolution methods
However, no technique is ever perfect so it does come with some downsides:
- ExM is incompatible with live cell imaging
- The more the sample is expanded, the more there is to image. This can mean more tiles, more Z-stacks, larger file sizes, longer times to analyze data and more time needed on the microscope
- Casting in a gel quenches fluorescence (depending on gel chemistry)
- Expanding reduces fluorophore density which lowers signal
- For deep imaging, long working distance objectives have a lower NA which reduces light collection efficiency, reducing signal
Current And Future Directions
Neuroscience
One of the key applications for ExM is in neuroscience imaging because there are still a lot of open questions surrounding the understanding of molecules and wiring in neural circuits.
ExM is a natural fit for this research because high resolution is required to identify key biological structures. It’s also typically performed using brain tissue slices which are ideal samples to be cast in a gel as they’re usually already fixed and the clearing process of expansion makes identification and mapping of the structures much easier (Karagiannis et al. 2018).
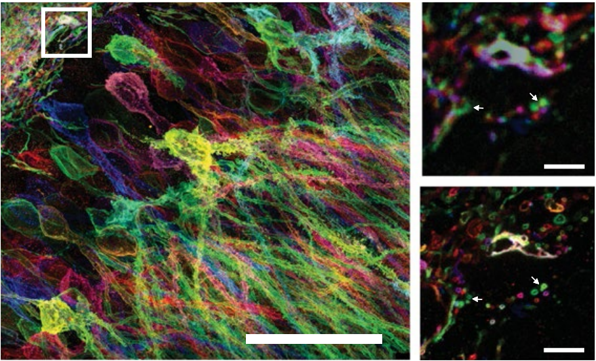
Adapted from Wassie et al. (2018)
RNA Imaging
Despite starting as a method to study proteins, recently developed methods allows ExM to also be used for RNA (Chen et al. 2016).
This allows fluorescence in situ hybridization (FISH) labeling strategies to be used to locate and identify RNA molecules, a technique called ExFISH. Aside from using this to image the nanoscale organization of RNA, the expansion decrowds RNA molecules which improves the quantification of RNA counts.
Signal amplification strategies frequently employed with FISH can also be used with ExFISH to increase the signal level for easy detection. This allows RNA to be detected at a single molecule level even in thick samples, answering questions relating the structure and location of RNA to biological functions.

Adapted from Wassie et al. 2018
Sequencing
Taking the mapping of RNA location a step further, it’s also possible to then use ExM to perform targeted or untargeted in situ sequencing of the RNA. One of the reasons this is possible is because of the decrowding of RNA molecules which facilitates access for the chemicals needed to make sequencing work. If RNA can be sequenced, the positions of specific genes can be identified, and patterns of gene expression revealed. This is extremely valuable in neuroscience for building up nanoscale-resolution maps of RNA in neurons or for identifying the organization of tumor cells in cancer biopsies.
This wouldn’t be possible without the fairly new technique, fluorescent in situ sequencing (FISSEQ) which allows the location of multiple RNA molecules to be pinpointed and then to determine the sequence of bases that identify each one of them. Combining this technique with ExM – expansion sequencing (ExSeq) – has a lot of potential in future sequencing research which is yet to be fully realized (Alon et al. 2020)
Choosing A Camera For Expansion Microscopy
ExM is a rare technique that can’t afford to trade off on any of the key camera specifications – sensitivity, field of view, speed or resolution.
As stated previously, many aspects of ExM result in a lower fluorescence signal than the same non-expanded sample. Casting in a gel quenches fluorescence, expanding reduces fluorophore density and low NA objectives for deep sample penetration reduce light collection efficiency. For these reasons, a high quantum efficiency camera with low noise is essential to collect as much of the remaining signal as possible.
Additionally, the more the sample is expanded, the more there is to image. This means potentially more tiles, more Z-stacks and more time needed on the microscope. Using a camera with the largest field of view possible for your system will ensure that as much of the sample as possible is captured in a single frame.
With the size of an expanded sample, creating three-dimensional images using light sheet or confocal microscopy is extremely time consuming. Imaging at a fast frame rate will speed this process up so a high-speed camera will help to capture the whole sample in less time.
Finally, as a super-resolution technique, maximizing resolution is one of the key concerns so choosing a camera with a pixel size that best matches the chosen objective NA will result in higher resolution images.
Summary
Compared with other super-resolution techniques, expansion microscopy offers some unique advantages in fluorophore flexibility, microscope flexibility, 3D imaging of thick samples, speed and compatibility with other techniques allowing them to become super-resolution methods.
It is also a young technique that is still experiencing new developments. One potential development is in the combination of multiple protocols to allow imaging of RNA, DNA and proteins together which would be transformative for transcriptomics research.
However, the technique is not without its downsides and its incompatibility with live cell imaging is one of the largest.
It will be very exciting to see how applications develop as more people add ExM to their repertoire of imaging techniques. As a super-resolution technique with one of the lowest barriers to entry this should only be a matter of time.
Acknowledgments
We’d like to thank Richie Kohman for comments on the text.
References
Alon, S., Goodwin, D. R., Sinha, A., Wassie, A. T., Chen, F., Daugharthy, E. R., Bando, Y., Kajita, A., Xue, A. G., Marrett, K., Prior, R., Cui, Y., Payne, A. C., Yao, C. Suk, H., Wang, R., Yu, C., Tillberg, P., Reginato, P., Pak, N., Liu, S., Punthambaker, S., Iyer, E. P. R., Kohman, R. E., Miller, J. A., Lein, E. S., Lako, A., Cullen, N., Rodig, S., Helvie, K., Abravanel, D. L., Wagle, N., Johnson, B. E., Klughammer, J., Slyper, M., Waldman, J., Jané-Valbuena, J., Rozenblatt-Rosen, O., Regev, A., Church, G. M. & Marblestone, A. H. (2020) Expansion Sequencing: Spatially Precise In Situ Transcriptomics in Intact Biological Systems. Science 29 Jan 2021:Vol. 371, Issue 6528, eaax2656 DOI: 10.1126/science.aax2656
Chen, F., Tillberg, P. W. & Boyden, E. S. (2015) Expansion microscopy. Science Jan 30; 347(6221): 543–548.
Chen, F. et al. (2016) Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684.
Chen, K., Ying-Chun, Z. & Guan-Fang, Z. (2019) Expansion Microscopy Imaging Technique and Its Application. Chinese Journal Of Analytical Chemistry Volume 47, Issue 5.
Gambarotto, D., Zwettler, F. U., Le Guennec, M., Schmidt-Cernohorska, M., Fortun, D., Borgers, S., Heine, J., Schloetel, J., Reuss, M., Unser, M., Boyden, E. S., Sauer, M, Hamel, V. & Guichard, P. (2019) Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nature Methods Volume 16, pages 71–74
Karagiannis, E. D. & Boyden, E. S. (2018) Expansion microscopy: development and neuroscience applications. Curr. Opin. Neurobiol. Jun;50:56-63. doi:10.1016/j.conb.2017.12.012.
Tillberg, P. W. & Chen, F. (2019) Expansion Microscopy:Scalable and Convenient Super-Resolution Microscopy. Annu. Rev. Cell Dev. Biol. Oct 6;35:683-701.
Wassie, A. T., Zhao, Y. & Boyden, E. S. (2018) Expansion microscopy: principles and uses in biological research. Nature Methods Volume 16, pages 33–41
Yu, C. J., Barry, N. C., Wassie, A. T., Sinha, A., Bhattacharya, A., Asano, S., Zhang, C., Chen, F., Hobert, O., Goodman, M. B., Haspel, G., Boyden, E. S. (2020) Expansion microscopy of C. elegans. eLife 9:e46249.
