Introduction
To support our claim that the Prime 95B is the best camera for localization microscopy, we have collaborated with some of our customers to compare the localization accuracy of the Prime 95B to popular EMCCD cameras.
We also investigated the improvement made by PrimeEnhance™, the live denoising algorithm that accompanies the Prime 95B.
To perform these tests, DNA-origami nanostructures with a pre-defined pattern of fluorophores were provided by GATTAquant and used as precision standards.
Prime 95B vs 1024×1024 EMCCD
Tests were performed by the Shim group at Korea University who compared the Prime 95B with PrimeEnhance™ to a 1024×1024 EMCCD. They used GattaPAINT80 surface-immobilized DNA origami nanostructures labeled with Atto655 with three localization sites per structure spaced 80 nm apart.
They excited the nanostructures with 647 nm laser excitation with an approximate intensity of 2kW/cm2 and a 20 ms exposure time over 30000 frames in TIR illumination.


The calculated mean localization accuracy of the 1024×1024 EMCCD was 13.29 nm compared to the Prime 95B which was 9.24 nm, providing a 30% increase in localization accuracy. The mode of the localization accuracy also showed a considerable difference, 11 nm compared to 6 nm.
These results show that the Prime 95B provides a significant increase in localization accuracy over the 1024×1024 EMCCD camera. The group then investigated PrimeEnhance™.
Prime 95B with PrimeEnhance™

It is important to note that enabling PrimeEnhance™ dropped the acquisition speed by ~40% with a 20 ms exposure time, but this discrepancy can be eliminated by using a 100 ms exposure time.
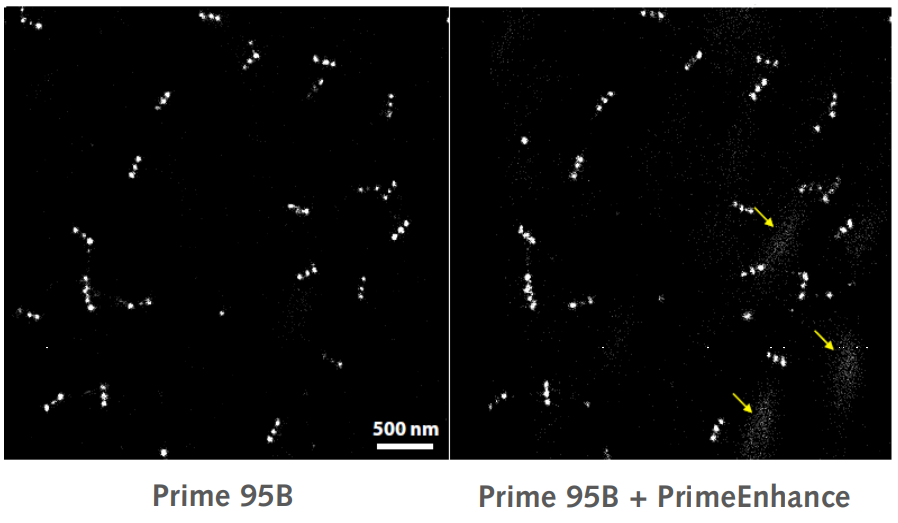
Initial results showed some noise signals in the localization image (marked by yellow arrows) when using PrimeEnhance™ . The group was able to remove these signals by increasing the intensity threshold in the analysis software by a factor of two.

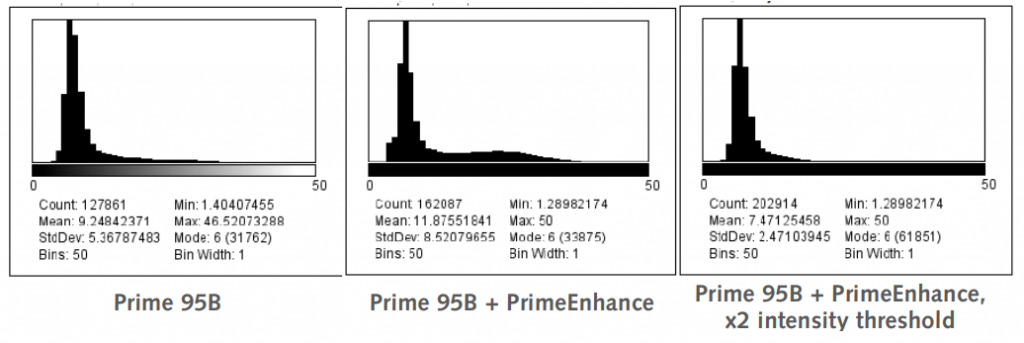
The histogram for Prime 95B with PrimeEnhance™ also showed some artifacts in the shape of an elongated tail which was noticeably removed when increasing the intensity threshold by a factor of two. The resulting mean localization accuracy increased from 9.24 nm on the Prime 95B to 7.47 nm, providing a further 20% increase in localization accuracy when using PrimeEnhance™ with the x2 intensity threshold. This shows that PrimeEnhance™ can give even higher quality data than the Prime 95B alone.
Prime 95B vs 512×512 EMCCD
Further tests were performed with another customer who compared the Prime 95B to a 512×512 EMCCD.
This test used Gatta-STORM 50R surface-immobilized nanorulers labeled with Alexa647 with two localization sites per structure spaced 50 nm apart. The nanostructures were excited with a 635 nm laser at a 100 ms exposure time over 10000 frames in TIRF illumination.
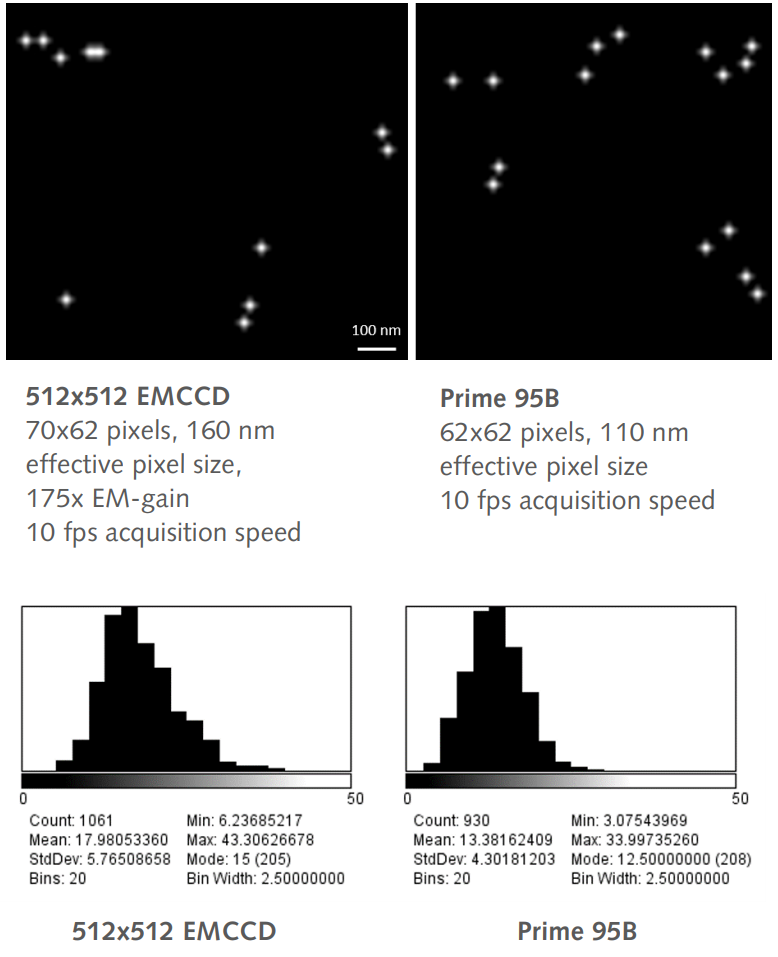
The calculated mean localization accuracy of the 512×512 EMCCD was 17.98 nm compared to the 95B which was 13.38 nm, providing a 25% increase in localization accuracy. The mode of the localization accuracy also showed a considerable difference, 15 nm compared to 12.5 nm.
Summary
The Prime 95B is the perfect solution for super-resolution localization microscopy.
We would recommend, based on this data and data from many customers, that the Prime 95B with its speed, field of view and sensitivity outperforms EMCCD for all the cases we have seen. Combined with this, PrimeEnhance™ may give an extra advantage for improving localization accuracy.
In addition, the camera’s PrimeLocate™ feature helps further by reducing the data acquired per frame, allowing for quicker analysis, reducing storage space, and allowing easier transport of data between computers/networks.
Download As PDF
References
We would like to thank the following companies and labs for their part in producing this data:
GATTAquant (http://www.gattaquant.com/) for providing the DNA-origami nanostructures
Abbelight (https://www.abbelight.com/) for providing the imaging buffer.
ThunderSTORM (https://zitmen.github.io/thunderstorm/) which was used to reconstruct and visualize the data.
The Shim group at Korea University (http://sodaus.wixsite.com/shimku).
