Introduction
Some super-resolution data cannot be visualized directly, only after images are reconstructed and processed can they be displayed, such as in PALM and STORM. This processing requires specialized algorithms and analysis software, which must be capable of dealing with the large datasets of thousands of frames from 3D, fast-moving dynamic samples with hundreds of data points of varying densities.
This software processing can also apply to single-molecule localization microscopy (SMLM), where analysis can be vital in order to optimize data output. Choices made in both pre- and post-processing make a big difference to the image output quality, changing important factors such as the signal-to-noise ratio, detection accuracy, and subpixel fluorophore localization. There are more than 30 software packages used for SMLM by the community, each varying in accessibility and processing power which have been subjected to quantitative analysis by studies such as the one by Sage and colleagues (2015). These software typically excel at analysis when individual fluorophores are separated by a distance >1.5 µm, but can struggle with densely-packed samples where fluorophores can be within 1 µm adjacent, with artifacts such as decreased precision, false positives, and missed detections arising. This could be solved by ensuring samples remain at a certain density, but especially in the case of living, dynamic biological samples this is not always possible and can change from frame to frame.
One solution to this issue is super-resolution optical fluctuation imaging (SOFI), but SOFI is a MATLAB package and can be challenging to use, has a non-linear response to brightness in higher-order modes, and doesn’t take temporal fluctuations into consideration (only using the local spatial information). Another analytical approach is super-resolution radial fluctuations or SRRF (pronounced ‘surf’), developed by Henriques and colleagues in 2016.
Principle of SRRF
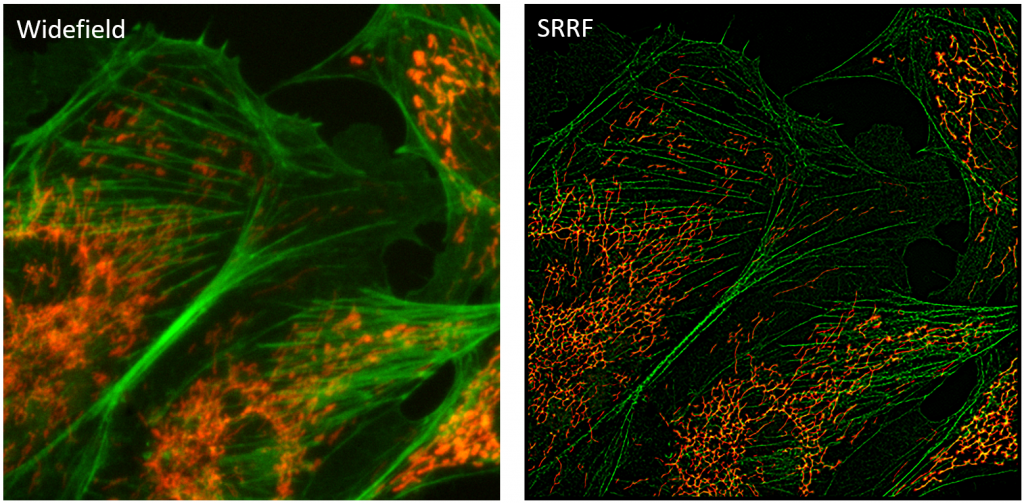
SRRF is a super-resolution algorithm that analyses a sequence of images to directly generate a super‑resolution image, without the need for fluorophore detection and localization. This results in spatial resolutions approaching ~60 nm and temporal resolutions of 1 second from live samples imaged with conventional fluorophores, using low-intensity illumination and acquiring as few as 100 frames.
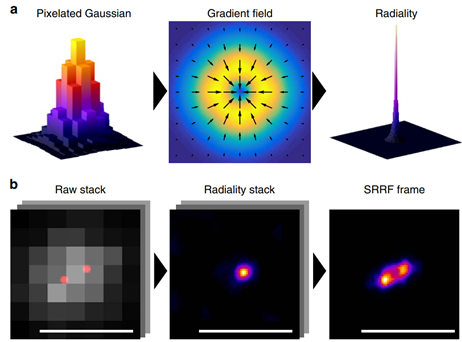
As described in the name, SRRF functions by observing fluctuations in radial symmetry throughout the image frames. The assumption is that the point spread function (PSF) of fluorescent molecules have a higher degree of radial symmetry than the background. SRRF magnifies each pixel into subpixels, and each is given a value related to the probability of it containing a fluorophore. This probability is based on the radial symmetry or ‘radiality’ that fluorophore PSFs have, due to being circular in nature. Radiality is measured for each subpixel in the image by taking a ring of neighboring subpixels and measuring the convergence of intensity gradients passing through these. Subpixels closer to the true center of molecules have higher radiality values, whereas subpixels far from molecule centers have lower radiality values (Fig.2 top row).
This calculation of the degree of radial symmetry is performed across each frame in the dataset on a sub-pixel basis. This compresses the effective PSF within the image to be sharper and of higher resolution than the original microscope PSF (Fig. 3a). Combining this with temporal analysis allows for further de-noising, as noise-induced peaks do not typically repeat over time and can be suppressed, and higher-order temporal analysis increased resolution further (akin to SOFI). This analysis generates a single super-resolution frame, and the process is highlighted in Fig.1.
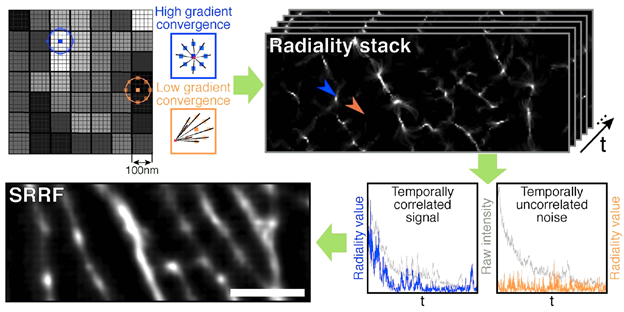
While radiality analysis comprises spatial information, signal fluctuations in time translate to radiality fluctuations in time, and so by performing temporal analysis it is clear if certain points are noise or signal. This temporal correlation analysis is similar to SOFI, and by combining it with spatial radiality information, SRRF presents a powerful platform for super-resolution analysis of samples.
The same group has since developed another algorithm termed SQUIRREL (super-resolution image rating and reporting of error locations), which can assess the quality of images produced with any super-resolution analysis method under different conditions, including SRRF (Culley et al. 2018a).
Advantages of SRRF
If fluorophores overlap by a distance smaller than the PSF width, this would be solved in techniques like STORM and PALM by ensuring that individual fluorophores appear independently of one another in different frames. However, if fluorophores persistently overlap then they cannot be successfully identified as separate molecules in traditional SMLM analysis, and may be lost from the analysis entirely. SRRF somewhat avoids this problem by (instead of trying to localize individual molecules), creating probability maps of where molecules are likely to be based on their radiality and temporal properties. Due to this, SRRF is suited to the analysis of samples that feature ultra-high densities (a high degree of overlap), which are typically more difficult to analyze with other software packages.
STORM and PALM super-resolution techniques can rely on high laser intensities in order to illuminate samples, receive brighter signals (for easier localization), and favorable blinking statistics. This can be limiting with live-cell work, as samples can become bleached and phototoxicity can build up. As SRRF does not rely on the distinct fluorophore blinking events required for STORM/PALM, lower laser intensities (which correspondingly result in more overlapping fluorophores). SRRF has performed better with some live samples when compared to other super-resolution techniques such as STED (Retzer et al. 2017).
SRRF also allows for a big resolution upgrade for typical widefield systems and TIRF systems, with the latter allowing for the combination of SRRF and TIRF, where the resolution improved from ~330 nm to ~108 nm resolution (Culley et al. 2018).
Scientific Cameras for SRRF
For super-resolution techniques such as SRRF, the choice of scientific camera can make a big difference. With exposure time being a factor that can affect SRRF image quality, it is vital to use a camera that is highly sensitive in low-light applications, allowing for sufficient signal detection even with lower exposure times. This can be achieved with high quantum efficiency devices such as modern back-illuminated sCMOS cameras.
It should be noted that some sCMOS cameras can feature a split sensor and fixed pattern (column) noise, this can be a challenge for SRRF as the algorithm can amplify these fixed patterns. Back-illuminated sCMOS cameras that use a single sensor and a clean, pattern-free bias are better for SRRF analysis.
Camera pixel size is also an important factor to consider with super-resolution. This can be a limiting factor for EMCCD technologies, which tend to use larger pixels to promote sensitivity at the cost of resolution. Back-illuminated sCMOS cameras typically feature smaller pixels balanced for Nyquist at high magnifications, allowing for optimized resolution which retaining a high sensitivity, larger FOV, and higher speed.
Summary
SRRF is a unique super-resolution algorithm that allows for fast live-cell imaging with standard fluorescent labels, even when there is an ultra-high density of emitters within an image. SRRF allows for cell biology experiments to upgrade to super-resolution by making use of an elegant analysis method where images are processed based on their radial symmetry. SRRF can be combined with other applications such as TIRF or light sheet, along with large sensitive sCMOS detectors in order to make super-resolution imaging more accessible.
Acknowledgments
Many thanks to Dr. Siân Culley for consulting on this article.
References
Culley, S., Albrecht, D., Jacobs, C., Pereira, P. M., Leterrier, C., Mercer, J., & Henriques, R. (2018a). Quantitative mapping and minimization of super-resolution optical imaging artifacts. Nature methods, 15(4), 263–266. https://doi.org/10.1038/nmeth.4605
Culley, S., Tosheva K.L., Pereira, P. M., & Henriques, R. (2018b) SRRF: Universal live-cell super-resolution microscopy, Intl J.Bio.CellBio, 101 74-79 https://doi.org/10.1016/j.biocel.2018.05.014.
Gustafsson N., Culley S., Ashdown G., Owen D.M., Pereira P.M., Henriques R. (2016) Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations, Nature Communications, 7, 12471, DOI: 10.1038/ncomms12471
Sage, D., Kirshner, H., Pengo, T., Stuurman, N., Min, J., Manley, S., & Unser, M. (2015). Quantitative evaluation of software packages for single-molecule localization microscopy, Nature methods, 12(8), 717–724. https://doi.org/10.1038/nmeth.3442
Retzer, K., Lacek, J., Skokan, R., Del Genio, C. I., Vosolsobě, S., Laňková, M., Malínská, K., Konstantinova, N., Zažímalová, E., Napier, R. M., Petrášek, J., & Luschnig, C. (2017). Evolutionary Conserved Cysteines Function as cis-Acting Regulators of Arabidopsis PIN-FORMED 2 Distribution. International journal of molecular sciences, 18(11), 2274. https://doi.org/10.3390/ijms18112274
