Jonathan Jeffet, Prof. Yuval Ebenstein
NanoBioPhotonix Lab, Department of Chemical Physics, Tel-Aviv University, Israel
Background
The NanoBioPhotonix Lab of Tel-Aviv University is led by Prof. Ebenstein and focuses on single-molecule genomics research and development of novel imaging techniques. Jonathan Jeffet is a PhD student and physicist from this highly multidisciplinary lab and works with their imaging systems performing optical genome mapping. This technique is complementary to typical DNA sequencing, where sequencing works by attempting to assemble short DNA segments, and optical mapping uses fluorescent tags with long fragments of DNA to create a ‘map’, this then helps to align sequences that would be problematic with other techniques.
As Mr Jeffet says: “We use fluorescence to tag DNA molecules, DNA damage, methylation, and we want to see all this genetic and epigenetic information on a single DNA molecule, this is the focus of the lab.”
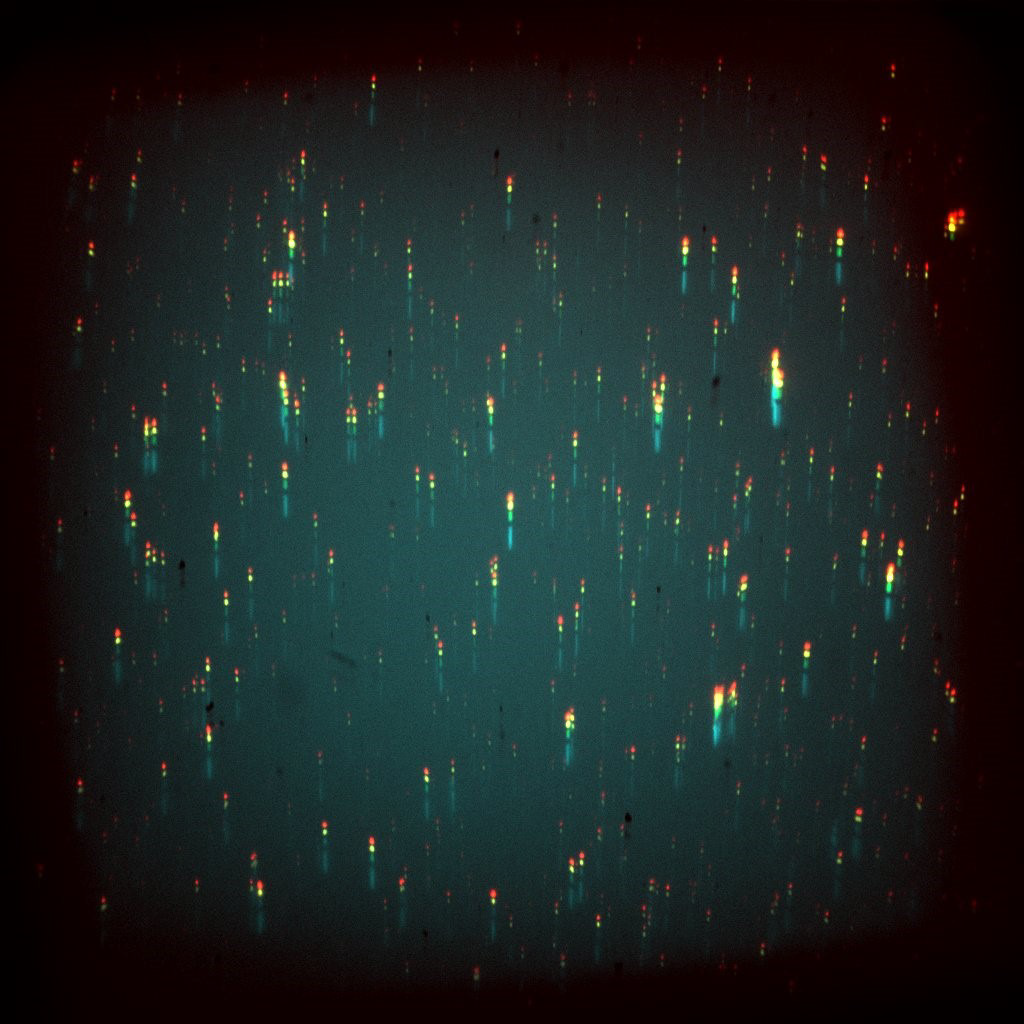
Challenge
From Mr. Jeffet: “We try to image all these genetic and epigenetic modifications on a single DNA molecule together, if we sequentially image different colors then we lose information as the molecules move around while we switch filters.”
The lab uses a five-laser microscope, with the five light sources ranging from 405-785 nm meaning the experiments are working with the entire visible spectrum to acquire images for optical genome maps, resulting in a complex system requiring advanced triggering and illumination methods. Sensitivity is required across the visible spectrum in order for experimental flexibility. In addition, the scale of the samples means a high resolution is needed, and due to the type of flat-field excitation used, only 60x magnification was available.
The markers are single-molecule fluorescent tags that have a very low signal, meaning that imaging at higher speeds is limited by the need for a good signal-to-noise ratio, with longer exposures needed to collect enough signal for a good image.
The [Prime BSI] was really quick and easy to set up and worked well with MicroManager, and we’re happy because we’re seeing better results, the SNR is really better, it’s perfect for our needs
Solution
The high speed, low noise, and high sensitivity of the Prime BSI resulted in a big increase in signal-to-noise ratio with the single-molecule imaging for the NanoBioPhotonix Lab. The 6.5 um pixel of the Prime BSI is optimized for Nyquist at 60x magnification, which results in high-resolution images.
The multiple modes of the Prime BSI also allow for flexible imaging, offering both a high dynamic range mode and a high sensitivity CMS mode, making the Prime BSI a solution for a wide range of different single-molecule experiments.
The Prime BSI was an improvement on previous CMOS solutions, as outlined by Mr. Jeffet: “We previously used a front-illuminated sCMOS and the SNR was much worse at the same acquisition rates… the [Prime BSI] allowed us to see more of the sample and our readings were much better with this camera.”
References
Jeffet J., Michaeli Y., Torchinsky D., Israel-Elgali I., Shomrom N., Craggs T.D., Ebenstein Y. (2020) Multi-modal Single-molecule Imaging with Continuously Controlled Spectral-resolution (CoCoS) Microscopy, bioRxiv 2020.10.13.330910; doi: https://doi.org/10.1101/2020.10.13.330910

