Assis. Prof. Matthew Lew
Department of Electrical & Systems Engineering, Washington University, Saint Louis
Background
The Lew lab is creating technology to image multidimensional trajectories of individual molecules in time and space, with one focus on understanding the structural dynamics of amyloid aggregation in diseases like Alzheimer’s.
In collaboration with Jan Bieschke, UCL, the lab has developed a single-molecule localization microscopy (SMLM) technique called TAB, transient amyloid binding, to image the structure of amyloid protein aggregates over periods of hours to days. Using a combination of engineered point spread functions (PSFs) such as Tri-spot and a new robust statistical estimation algorithm, they unmix molecular position and orientation within raw SMLM data, allowing them to follow translation and rotation (azimuthal and polar angles and wobble) of molecules over time.
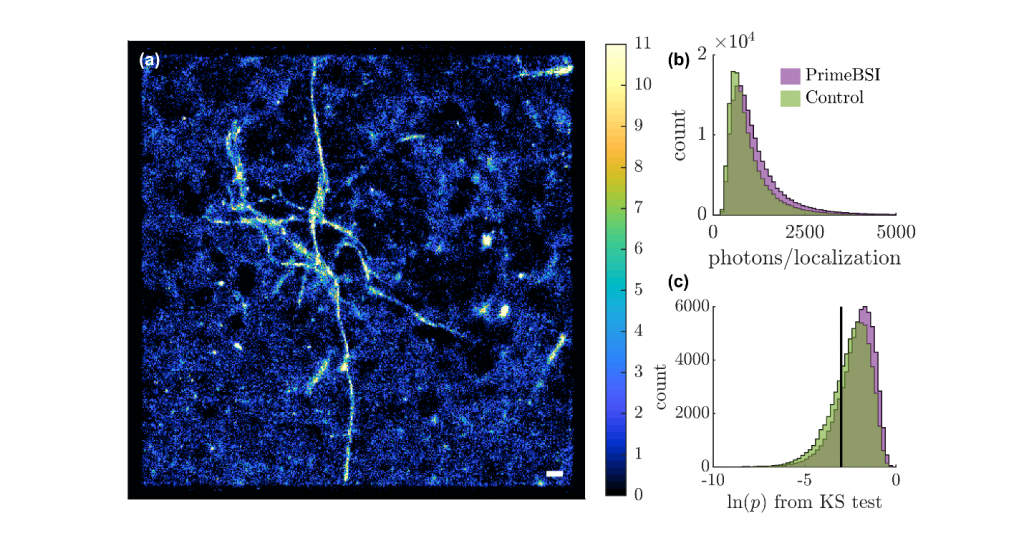
Challenge
Localizing individual molecules in super-resolution fluorescence microscopy is photon limited, with the precision improving by the inverse of the square root of the number of photons collected. Beyond standard SMLM, orientation-sensing SMLM (using the Tri-spot PSF), requires the fixed photon budget of a single molecule to be spread across a larger area of the detector.
With less than 1000 photons detected within each localization in TAB imaging, previous sCMOS cameras struggled to produce robust orientation resolved images due to suboptimal quantum efficiency and sensitivity. Further, pixel to pixel variability in electronic gain adds undesired noise to photon counts in each pixel, thereby polluting sensitive measurements of molecular orientation.
Ultimately, the best optical imaging comes from using the most accurate and sensitive photon-counting camera. The Prime BSI has enabled my lab to push the envelope of nanoscale imaging and develop new ways of seeing single molecules in living systems.
Solution
The Prime BSI’s high quantum efficiency, low read noise, and very low fixed pattern noise improves the precision of measuring molecular orientation using a variety of PSFs. The higher quantum efficiency enables more molecules to be detected and increases the photons collected from each localization. Reading out photon counts from the camera that match closely the Poisson statistical distribution ensures that the imaging achieves the best-possible shot noise limit. Having more localizations, each with better precision, creates super-resolved images with higher resolution and resolve heterogeneities in molecular orientations that could not be observed before.

